7. Multidimensional input¶
lsqfitgp supports multidimensional input through numpy structured
arrays. Elements of
structured arrays have named fields. Easier done than said:
import numpy as np
x = np.linspace(-3, 3, 30)
y = np.linspace(-3, 3, 30)
xy = np.empty((len(x), len(y)), dtype=[('x', float), ('y', float)])
xy['x'] = x[:, None]
xy['y'] = y[None, :]
Here we used a bit of Numpy indexing and broadcasting to fill a 30x30 array
xy with a grid of x, y coordinates. Let’s look at it in order. We specified
a data type [('x', float), ('y', float)] for the array xy, this means
that each element of xy is a pair of numbers. We specified a shape
(len(x), len(y)), where x and y are the two linspaces, and then
filled separately the 'x' and 'y' fields of the xy elements with
the linspaces. xy is a 2D array, while x and y are 1D, so using
None we added a dummy dimension along which the copying is repeated. The
colons : are empty slices and leave the dimension untouched.
It is not important for xy to be a 2D array because we are using
bidimensional points, it is just a convenience for building a rectangular grid.
Do not confuse the number of dimensions of the array with the number of fields
in its elements.
A thing to notice is that we only used 30 points for the side of the grid. This
is because 30x30 = 900 and at around 1000 datapoints lsqfitgp starts
being slow. This is an inherent limitation of Gaussian processes, which can be
only overcome with approximations or specialized algorithms. If you need to
handle a lot of datapoints, pymc3 has some support for separable
kernels on grids, while celerite implements harmonic oscillators
in 1D.
Now we put xy into a GP as usual and extract a sample from the
prior.
import lsqfitgp as lgp
import gvar
gp = lgp.GP(lgp.ExpQuad())
gp.addx(xy, 'foo')
prior = gp.prior('foo')
sample = gvar.sample(prior)
We plot the sample following the matplotlib 3d tutorial:
from matplotlib import pyplot as plt
fig, ax = plt.subplots(num='lsqfitgp example', subplot_kw=dict(projection='3d'))
ax.plot_surface(xy['x'], xy['y'], sample, cmap='viridis')
fig.savefig('in1.png')
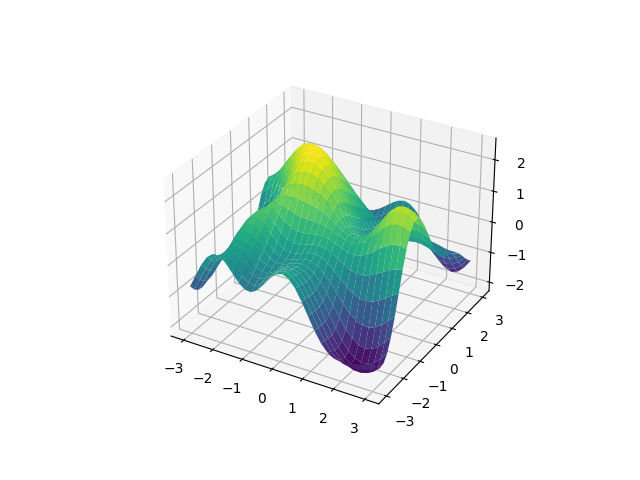
We got a nice random surface. The ExpQuad kernel just worked out of
the box with multidimensional input. What kernel are we using really?
ExpQuad is a subclass of IsotropicKernel. This means it only
depends on the distance between points, i.e., it doesn’t care what the input is
as long as it can apply Pythagoras’ theorem. So the kernel we used is:
We can do more sophisticated things if we control how the kernel acts on each dimension. Let’s use a random walk along \(x\) and an exponential quadratic along \(y\):
gp = lgp.GP(lgp.Wiener(dim='x', loc=-3) * lgp.ExpQuad(dim='y'))
gp.addx(xy, 'foo')
prior = gp.prior('foo')
sample = gvar.sample(prior)
ax.cla()
ax.plot_surface(xy['x'], xy['y'], sample, cmap='viridis')
fig.savefig('in2.png')
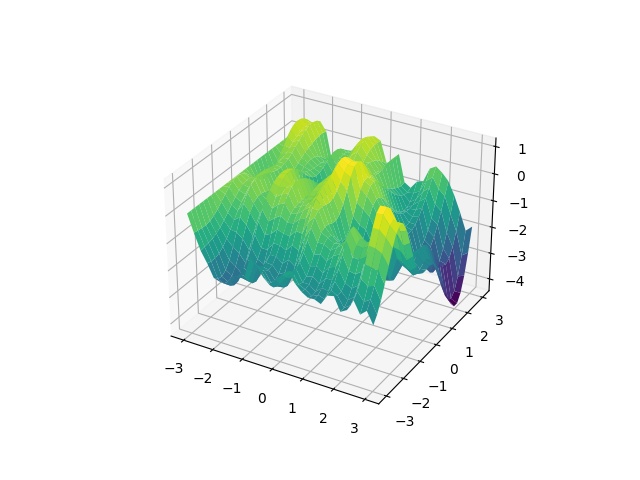
Uhm, quite unelegant. I don’t like random walks any more. We used the dim
keyword to specify the dimension, and loc=-3 to translate the random walk
(it raises an error if it gets negative input points).
In many dimensions it can get tedious to specify all the field names, it would
be simpler if multidimensional input was implemented with arrays. Luckily
numpy supports array data types in fields. Let’s redo the first example,
this time without named fields:
xy = np.empty((len(x), len(y)), dtype=[('foo', float, 2)])
xy['foo'][..., 0] = x[:, None]
xy['foo'][..., 1] = y[None, :]
Ok, I lied a bit, there’s still a field name around. It is needed because
otherwise lsqfitgp would not know that the last axis with size 2 is a
2-dimensional point instead of a pair of points, so we encapsulate it in a
field. numpy treats the ellipsis ... as a string of colons : of
the appropriate length.
gp = lgp.GP(lgp.ExpQuad())
gp.addx(xy, 'foo')
prior = gp.prior('foo')
sample = gvar.sample(prior)
ax.cla()
ax.plot_surface(xy['foo'][..., 0], xy['foo'][..., 1], sample, cmap='viridis')
fig.savefig('in3.png')
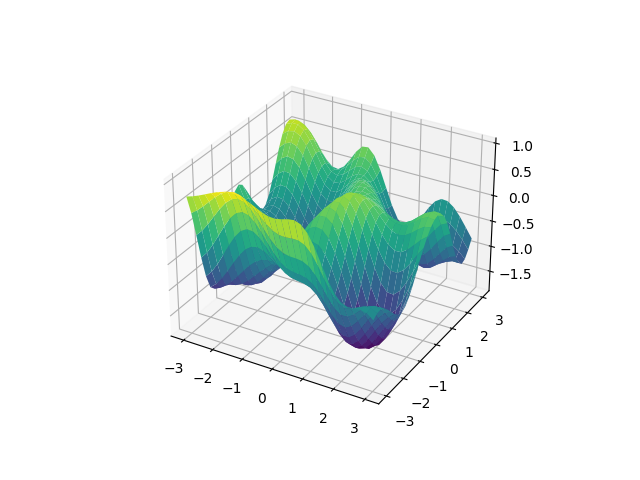
However, with this method, it is not possible to apply a kernel only along a specific dimension. A kernel can be applied only on everything or on a single whole named field. By splitting your dimensions in named fields with different shapes this should still be flexible enough, nevertheless in a future release I will introduce support for applying a kernel along any kind of subset of the dimensions.