10. Splitting components¶
In More on kernels we saw an example where we summed two ExpQuad
kernels with different scale, and the result effectively looked like the sum
of two processes, because the kernel of a sum of processes is the sum of their
kernels.
When summing processes it is useful to get the fit result separately for each
component. In lsqfitgp there’s not a specific tool for this because it
can be implemented using kernel tricks, multidimensional input and
transformations.
Let’s see. We first generate some data:
import numpy as np
import lsqfitgp as lgp
import gvar
from matplotlib import pyplot as plt
gp = lgp.GP(10 * lgp.ExpQuad(scale=10) + lgp.ExpQuad(scale=1))
x = np.linspace(-10, 10, 21)
gp.addx(x, 'pinguini')
prior = gp.prior('pinguini')
y = gvar.sample(prior)
fig, ax = plt.subplots(num='lsqfitgp example')
ax.plot(x, y, '.k')
fig.savefig('components1.png')
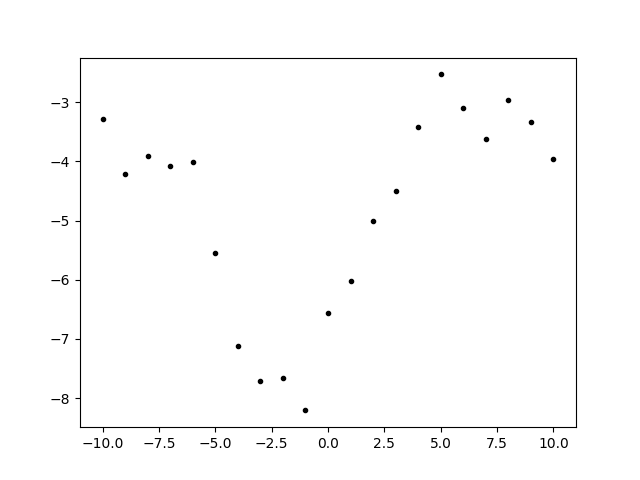
We made a mixture of two exponential quadratics and took a sample from the prior as data. Generating fake data from the prior is a good way to make sure that the fit will make sense with the data.
Now we setup a fit with an additional integer input dimension that indicates the component, similarly to what we did in Multidimensional output:
xtype = np.dtype([
('x' , float),
('comp', int ),
])
xcomp = np.empty((2, len(x)), xtype)
xcomp['x' ] = x # broadcasting aligns the last axes
xcomp['comp'] = np.array([0, 1])[:, None]
We need to make a kernel that changes based on the value in the 'comp'
field. We can do that by using a Rescaling that acts on 'comp':
kernel1 = 10 * lgp.ExpQuad(scale=10)
kernel2 = lgp.ExpQuad(scale=1)
kernel = kernel1 * lgp.Rescaling(stdfun=lambda x: x['comp'] == 0)
kernel += kernel2 * lgp.Rescaling(stdfun=lambda x: x['comp'] == 1)
gp = lgp.GP(kernel)
The boolean operation in the stdfun argument returns 0 or 1, and since
stdfun is called on both arguments of the kernel, each component is nonzero
only when acting on two points which have the same 'comp'.
Now we add separately the two components and sum them with
addtransf():
gp.addx(xcomp[0], 'longscale')
gp.addx(xcomp[1], 'shortscale')
gp.addtransf({'shortscale': 1, 'longscale': 1}, 'sum')
We can now proceed as usual to get the posterior on other points:
xplot = np.empty((2, 200), dtype=xtype)
xplot['x'] = np.linspace(-10, 10, xplot.shape[1])
xplot['comp'] = np.array([0, 1])[:, None]
gp.addx(xplot[0], 'longscale_plot')
gp.addx(xplot[1], 'shortscale_plot')
gp.addtransf({'longscale_plot': 1, 'shortscale_plot': 1}, 'sum_plot')
post = gp.predfromdata({'sum': y})
ax.cla()
for sample in gvar.raniter(post, 2):
line, = ax.plot(xplot[0]['x'], sample['sum_plot'])
color = line.get_color()
ax.plot(xplot[0]['x'], sample['longscale_plot'], '--', color=color)
ax.plot(xplot[0]['x'], sample['shortscale_plot'], ':', color=color)
ax.plot(x, y, '.k')
fig.savefig('components2.png')
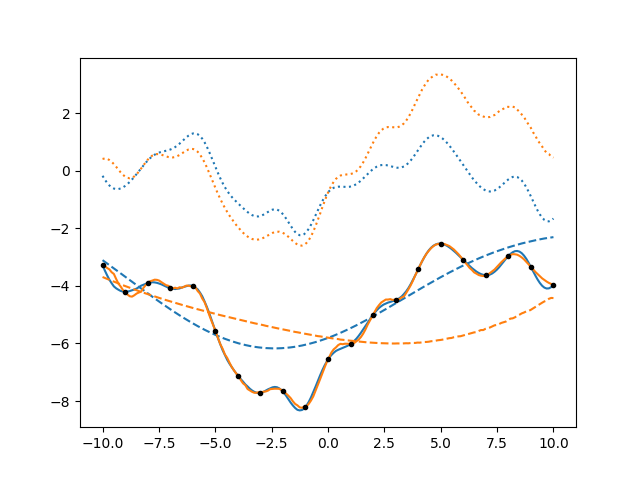
It’s interesting to note that the uncertainty on the individual components is larger than the uncertainty on the total, because there are various possible combinations that give the same data.
Writing this code was a bit tedious, I had to use Rescaling for each
kernel component, make a structured array and add separately the components,
and then redo it again for the plotting points. I’ll now rewrite the code in a
tidier way by defining a function and using where(). This time I’ll also
generate data separately for each component, although the fit will be done only
on the sum as before:
kernel = lgp.where(lambda x: x['comp'] == 0, kernel1, kernel2)
gp = lgp.GP(kernel)
keys = ['longscale', 'shortscale', 'sum']
def addxcomp(x, basekey):
xcomp = np.empty((2, len(x)), dtype=[('x', float), ('comp', int)])
xcomp['x'] = x
xcomp['comp'] = np.arange(2)[:, None]
gp.addx(xcomp[0], basekey + keys[0])
gp.addx(xcomp[1], basekey + keys[1])
gp.addtransf({
basekey + keys[0]: 1,
basekey + keys[1]: 1
}, basekey + keys[2])
x = np.linspace(-10, 10, 21)
xplot = np.linspace(-10, 10, 200)
addxcomp(x, 'data')
addxcomp(xplot, 'plot')
dataprior = gp.prior(['data' + k for k in keys])
y = gvar.sample(dataprior)
post = gp.predfromdata({
'datasum': y['datasum']
}, ['plot' + k for k in keys])
ax.cla()
for sample in gvar.raniter(post, 2):
line, = ax.plot(xplot, sample['plotsum'])
color = line.get_color()
ax.plot(xplot, sample['plotlongscale'], '--', color=color)
ax.plot(xplot, sample['plotshortscale'], ':', color=color)
for marker, key in zip(['x', '+', '.'], keys):
ax.plot(x, y['data' + key], color='black', marker=marker, label=key, linestyle='')
ax.legend()
fig.savefig('components3.png')
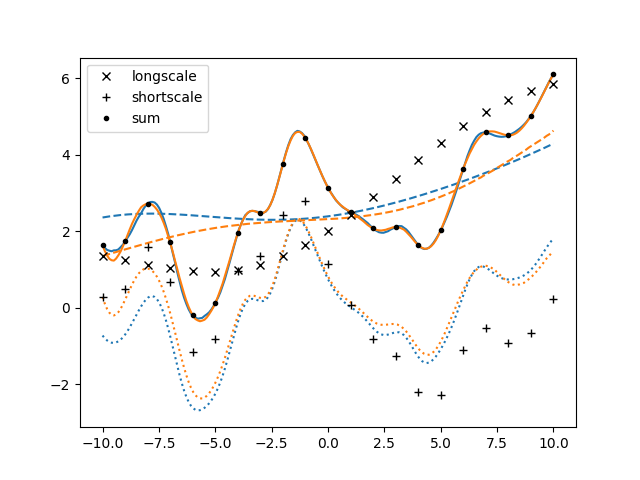
This version of the code was shorter and less redundant than the one we started
with, but it’s not very intuitive. We still have to take care manually of
indicating which component we are using by setting appropriately the 'comp'
field in the x arrays each time, and each time, for each set of points we
want to consider, we have to sum the two components after evaluating them
separately. There is another set of methods in GP designed to make
this kind of thing quicker. Let’s see. We start by creating a GP
object without specifying a kernel:
gp = lgp.GP()
Then we add separately the two kernels to the object using GP.addproc():
gp.addproc(kernel1, 'long')
gp.addproc(kernel2, 'short')
Now the two names 'long' and 'short' stand for independent processes
with their respective kernels. (These names reside in a namespace separate
from the one used by GP.addx() and GP.addtransf().) Now we use
these to define their sum as a process instead of summing them after
evaluation on specific points:
gp.addproctransf({'long': 1, 'short': 1}, 'sum')
The method GP.addproctransf() is analogous to GP.addtransf() but
works for the whole process at once. What we are doing mathematically is the
following:
Now that we have defined the components, we evaluate them on the points:
x = np.linspace(-10, 10, 21)
xplot = np.linspace(-10, 10, 200)
gp.addx(x, 'datalong' , proc='long' )
gp.addx(x, 'datashort', proc='short')
gp.addx(x, 'datasum' , proc='sum' )
gp.addx(xplot, 'plotlong' , proc='long' )
gp.addx(xplot, 'plotshort', proc='short')
gp.addx(xplot, 'plotsum' , proc='sum' )
We specified the processes using the proc parameter of GP.addx().
Then we continue as before:
dataprior = gp.prior(['datalong', 'datashort', 'datasum'])
y = gvar.sample(dataprior)
post = gp.predfromdata({
'datasum': y['datasum']
}, ['plotlong', 'plotshort', 'plotsum'])
ax.cla()
for sample in gvar.raniter(post, 2):
line, = ax.plot(xplot, sample['plotsum'])
color = line.get_color()
ax.plot(xplot, sample['plotlong'], '--', color=color)
ax.plot(xplot, sample['plotshort'], ':', color=color)
for marker, key in zip(['x', '+', '.'], ['long', 'short', 'sum']):
ax.plot(x, y['data' + key], color='black', marker=marker, label=key, linestyle='')
ax.legend()
fig.savefig('components4.png')
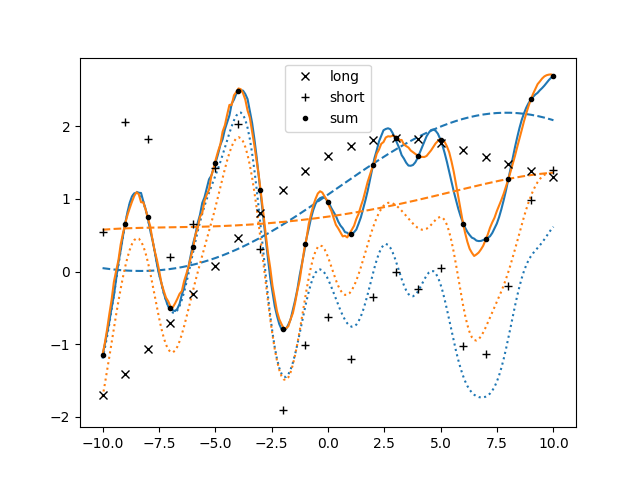
If there was this more convenient way of dealing with latent components, why
didn’t we introduce it right away? The reason is that it is not as general as
managing the components manually with an explicit field. GP.addproc()
defines processes which are independent of each other; to have nontrivial a
priori correlations it is necessary to put the process index in the domain such
that kernel can manipulate it. We did this at the end of Multidimensional output when
we introduced an anticorrelation between the random walk components by
multiplying the kernel with lgp.Categorical(dim='coord', cov=[[1, -0.99],
[-0.99, 1]]).