12. Hyperparameters in the input¶
This section is a bit technical, I will just explain a workaround necessary to
make empbayes_fit work when the hyperparameters enter in the
definition of the Gaussian process from the input points instead of from the
kernel parameters and the input points are multidimensional.
Let’s say you are investigating on a case of disappeared cats in a village. You want to study the correlation between the number of new cars bought per day and the number of reported disappeared cats per day.
To make it quick, I’ll first write the model and then generate the fake data from it for some value of the hyperparameters, then we’ll check if the fit recovers the correct hyperparameters.
import numpy as np
import gvar
import lsqfitgp as lgp
from matplotlib import pyplot as plt
def makepoints(time):
points = np.empty((2, len(time)), dtype=[
('time', float),
('series', int) # 0 = cats, 1 = cars
])
points['time'] = time
points['series'] = np.arange(2)[:, None]
return points
time = np.arange(20)
def makegp(hp):
delay = hp['delay']
corr = hp['corr']
scale = hp['scale']
kernel = lgp.ExpQuad(dim='time', scale=scale)
cov = np.array([[1, corr], [corr, 1]])
kernel *= lgp.Categorical(dim='series', cov=cov)
gp = lgp.GP(kernel)
points = makepoints(time)
points['time'][0] -= delay
gp.addx(points, 'data')
return gp
truehp = {
'delay': 10,
'corr': 0.70,
'scale': 3
}
gp = makegp(truehp)
prior = gp.prior('data')
data = gvar.sample(prior)
fig, ax = plt.subplots(num='lsqfitgp example')
ax.plot(time, data[0], '.k', label='cats')
ax.plot(time, data[1], 'xk', label='cars')
ax.legend()
fig.savefig('hyperstruct1.png')
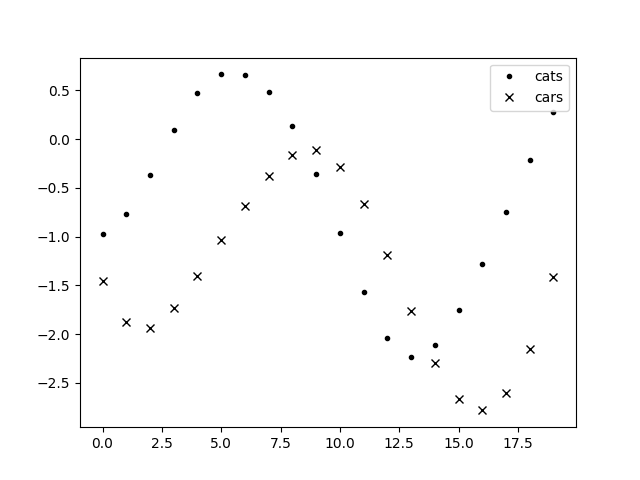
You can see that cats seem to reproduce the trend of cars but with a delay. Let’s fit now:
hprior = {
'delay': gvar.gvar(10, 5),
'func(corr)': gvar.BufferDict.uniform('func', -1, 1),
'log(scale)': gvar.log(gvar.gvar(3, 1))
}
fit = lgp.empbayes_fit(hprior, makegp, {'data': data})
hp = fit.p
When I run this last piece of code, it fails with:
ValueError: setting an array element with a sequence.
Ok, what went wrong? First, I used np.array() in makegp without doing
from autograd import numpy. Second, I did points['time'][0] -= delay,
which is an inplace operation, which is not allowed by autograd. Fixing the
first is easy, but how do we fix the second? There isn’t an obvious way to put
values into a numpy structured array without assigning to it. For this reason,
lsqfitgp provides a wrapper, StructuredArray, that allows
assigning to fields without breaking autograd:
from autograd import numpy as np
def makegp(hp):
delay = hp['delay']
corr = hp['corr']
scale = hp['scale']
kernel = lgp.ExpQuad(dim='time', scale=scale)
cov = np.array([[1, corr], [corr, 1]])
kernel *= lgp.Categorical(dim='series', cov=cov)
gp = lgp.GP(kernel)
points = makepoints(time)
points = lgp.StructuredArray(points)
points['time'] = np.array([time - delay, time])
# a StructuredArray can be assigned only a whole field at once
gp.addx(points, 'data')
return gp
fit = lgp.empbayes_fit(hprior, makegp, {'data': data}, raises=False)
# we use raises=False because the minimizer is a bit picky.
hp = fit.p
for k in truehp:
print(k, truehp[k], hp[k])
Output:
delay 10 10.57(28)
corr 0.7 0.66(14)
scale 3 2.993(36)
It seems to work. Let’s plot some samples:
timeplot = np.linspace(0, 19, 200)
xplot = makepoints(timeplot)
for hpsamp in gvar.raniter(hp, 2):
gp = makegp(hpsamp)
xplot[0]['time'] = timeplot - hpsamp['delay']
gp.addx(xplot, 'plot')
yplot = gp.predfromdata({'data': data}, 'plot')
for sample in gvar.raniter(yplot, 1):
ax.plot(timeplot, sample[0], color='blue', alpha=0.5)
ax.plot(timeplot, sample[1], color='red', alpha=0.5)
fig.savefig('hyperstruct2.png')
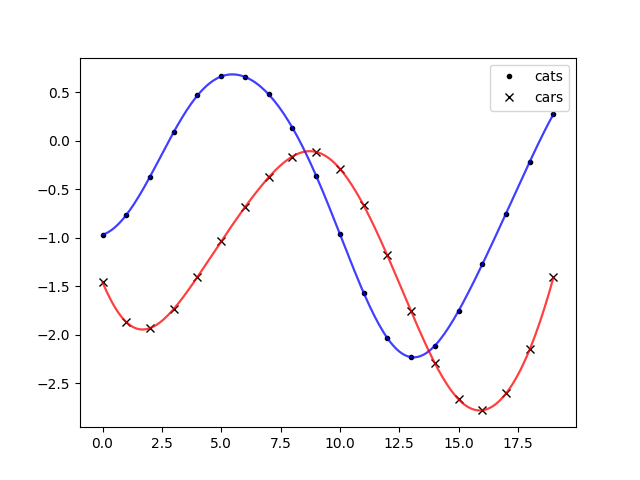
For better tidiness we should have put subtracting the delay from the time array for cats into the function makepoints.